Quickstart
Installation
Install phytest using pip:
pip install phytest
Quick Start
Phytest is a tool for automating quality control checks on sequence, tree and metadata files during phylogenetic analyses. Phytest ensures that phylogenetic analyses meet user-defined quality control tests.
Here we will create example data files to run our tests on.
Create an alignment fasta file example.fasta
>Sequence_A
ATGAGATCCCCGATAGCGAGCTAGCGATCGCAGCGACTCAGCAGCTACAGCGCAGAGGAGAGAGAGGCCCCTATTTACTAGAGCTCCAGATATAGNNTAG
>Sequence_B
ATGAGATCCCCGATAGCGAGCTAGXGATCGCAGCGACTCAGCAGCTACAGCGCAGAGGAGAGAGAGGCCCCTATTTACTAGAGCTCCAGATATAGNNTAG
>Sequence_C
ATGAGA--CCCGATAGCGAGCTAGCGATCGCAGCGACTCAGCAGCTACAGCGCAGAGGAGAGAGAGGCCCCTATTTACTAGAGCTCCAGATATAGNNTAG
>Sequence_D
ATGAGATCCCCGATAGCGAGCTAGCGATNNNNNNNNNNNNNNNNNTACAGCGCAGAGGAGAGAGAGGCCCCTATTTACTAGAGCTCCAGATATAGNNTAG
Create a tree newick file example.tree
(Sequence_A:1,Sequence_B:0.2,(Sequence_C:0.3,Sequence_D:0.4):0.5);
Writing a test file
- We want to enforce the follow constraints on our data:
The alignment has 4 sequences
The sequences have a length of 100
The sequences only contains the characters A, T, G, C, N and -
The sequences are allowed to only contain single base deletions
The longest stretch of Ns is 10
The tree has 4 tips
The tree is bifurcating
The alignment and tree have the same names
All internal branches are longer than the given threshold
There are no outlier branches in the tree
We can write these tests in a python files example.py
from phytest import Alignment, Sequence, Tree
def test_alignment_has_4_sequences(alignment: Alignment):
alignment.assert_length(4)
def test_alignment_has_a_width_of_100(alignment: Alignment):
alignment.assert_width(100)
def test_sequences_only_contains_the_characters(sequence: Sequence):
sequence.assert_valid_alphabet(alphabet="ATGCN-")
def test_single_base_deletions(sequence: Sequence):
sequence.assert_longest_stretch_gaps(max=1)
def test_longest_stretch_of_Ns_is_10(sequence: Sequence):
sequence.assert_longest_stretch_Ns(max=10)
def test_tree_has_4_tips(tree: Tree):
tree.assert_number_of_tips(4)
def test_tree_is_bifurcating(tree: Tree):
tree.assert_is_bifurcating()
def test_aln_tree_match_names(alignment: Alignment, tree: Tree):
aln_names = [i.name for i in alignment]
tree.assert_tip_names(aln_names)
def test_all_internal_branches_lengths_above_threshold(tree: Tree, threshold=1e-4):
tree.assert_internal_branch_lengths(min=threshold)
def test_outlier_branches(tree: Tree):
# Here we create a custom function to detect outliers
import statistics
tips = tree.get_terminals()
branch_lengths = [t.branch_length for t in tips]
cut_off = statistics.mean(branch_lengths) + statistics.stdev(branch_lengths)
for tip in tips:
assert tip.branch_length < cut_off, f"Outlier tip '{tip.name}' (branch length = {tip.branch_length})!"
Running Phytest
We can then run these tests on our data with phytest:
phytest examples/example.py -s examples/data/example.fasta -t examples/data/example.tree
Generate a report by adding --report report.html.
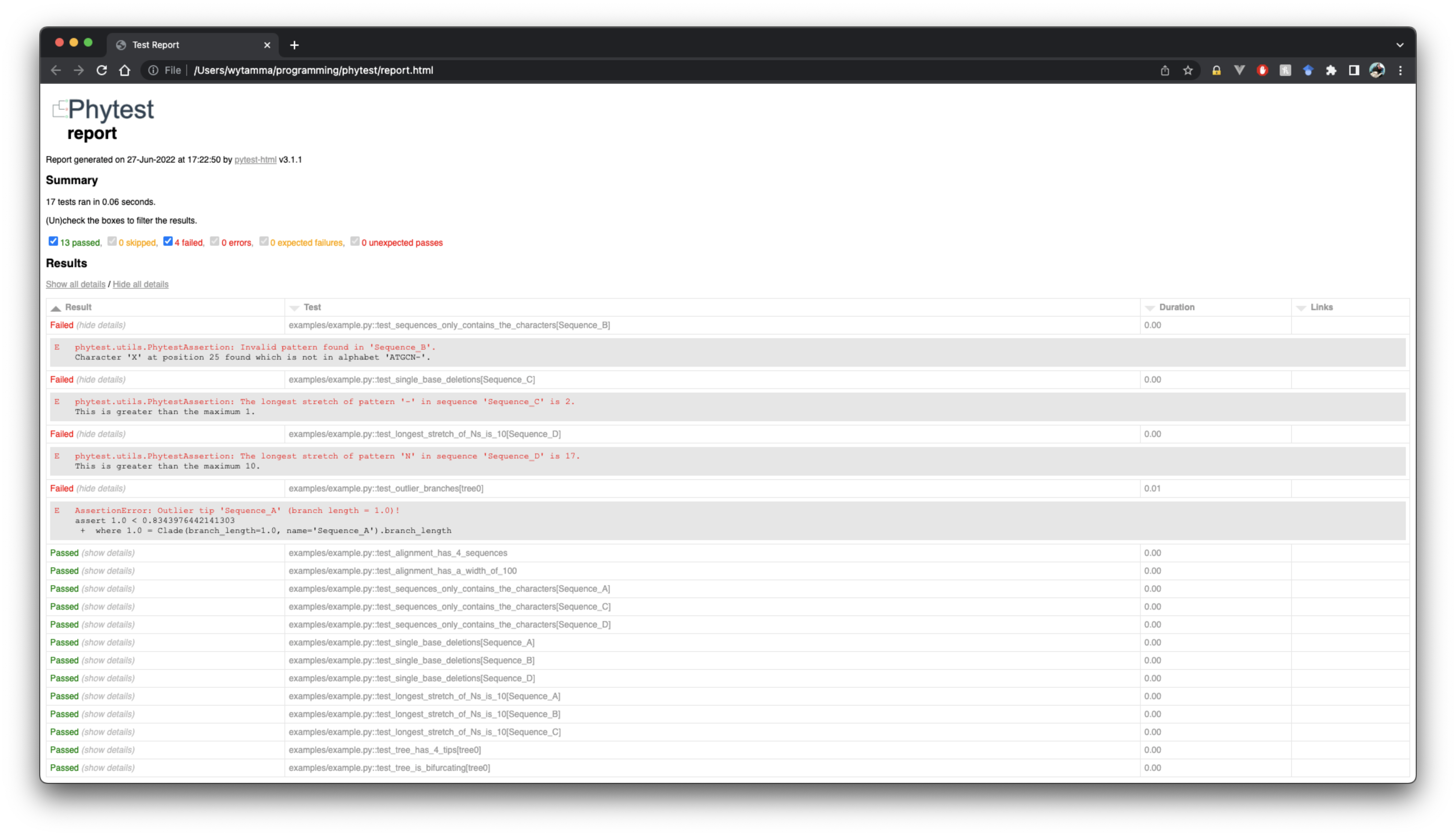
From the output we can see several tests failed:
FAILED examples/example.py::test_sequences_only_contains_the_characters[Sequence_B] - AssertionError: Invalid pattern found in 'Sequence_B'!
FAILED examples/example.py::test_single_base_deletions[Sequence_C] - AssertionError: Longest stretch of '-' in 'Sequence_C' > 1!
FAILED examples/example.py::test_longest_stretch_of_Ns_is_10[Sequence_D] - AssertionError: Longest stretch of 'N' in 'Sequence_D' > 10!
FAILED examples/example.py::test_outlier_branches - AssertionError: Outlier tip 'Sequence_A' (branch length = 1.0)!
Results (0.07s):
15 passed
4 failed
- examples/example.py:12 test_sequences_only_contains_the_characters[Sequence_B]
- examples/example.py:16 test_single_base_deletions[Sequence_C]
- examples/example.py:20 test_longest_stretch_of_Ns_is_10[Sequence_D]
- examples/example.py:32 test_outlier_branches